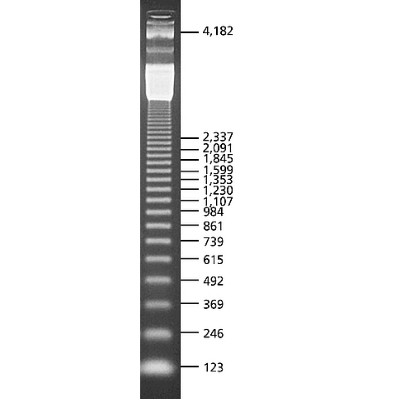
DNA Molecular Weight Marker III, DIG-labeled
DNA Molecular Weight Marker III, DIG-labeled is a fragment mixture prepared by cleavage of λDNA with EcoR I and Hind III. The size ranges from 0.12 to 21.2kbp. A photodigoxigenin has been introduced at approx. every 200th to 300th base in the DNA fragments.
We are committed to bringing you Greener Alternative Products, which adhere to one or more of The 12 Principles of Greener Chemistry. This product is designed as a safer chemical. The DIG System was established as a sensitive and cost-effective alternative to radioactivity for the labeling and detection of nucleic acids. There are many available publications that prove the versatility of the DIG System, so use of radio-labeling is no longer the only option for labeling of DNA for hybridization.
DNA Molecular Weight Marker III, DIG-labeled, has been used as a size standard in Southern blot analysis for nucleic acid labeling and detection.[1]
We are committed to bringing you Greener Alternative Products, which adhere to one or more of The 12 Principles of Greener Chemistry. This product is designed as a safer chemical. The DIG System was established as a sensitive and cost-effective alternative to using radioactivity for the labeling and detection of nucleic acids. There are many available publications that prove the versatility of the DIG System, so use of radio-labeling is no longer the only option for labeling of DNA for hybridization.
The mixture contains 13 fragments wit the following base pair lengths: 125, 564, 831, 947, 1375, 1584, 1904, 2027, 3530, 4268, 4973, 5148, and 21226 bp.
Note: Fragment lengths are derived from computer analysis of the DNA sequence.
Depending on the size range of the marker, the smallest fragments will only be visible on overloaded gels.
For life science research only. Not for use in diagnostic procedures.
Fragment lengths are derived from computer analysis of the DNA sequence. Depending on the size range of the marker the smallest fragments will only be visible on overloaded gels.